Our lab is in the Departments of Genetics and Neurology at Albert Einstein College of Medicine. We are located in the Michael F. Price Center for Genetics and Translational Medicine, and affiliated with the Department of Neuroscience, the Data Science Institute, the Rose F. Kennedy Center for Intellectual and Developmental Disabilities Research, and the Montefiore Einstein Comprehensive Cancer Center. The research areas of the lab are computational genomics and bioinformatics, with a strong focus on mining and interpreting large-scale genomic data (ie, bigdata science). The biological themes of our research are centered on the genetic and epigenetic regulations of early development, including neural development and heart development, and their dysregulation in diseases. We also apply our bioinformatics framework and expertise to study cancer development and treatment. We are always motivated by the continuous needs of innovative bioinformatics research to address new challenges in bigdata from evolving omics technologies. Our software and codes are publicly shared at the GitHub (github.com/bioinfoDZ), in support of research rigor, reproducibility and transparency.
Our Research
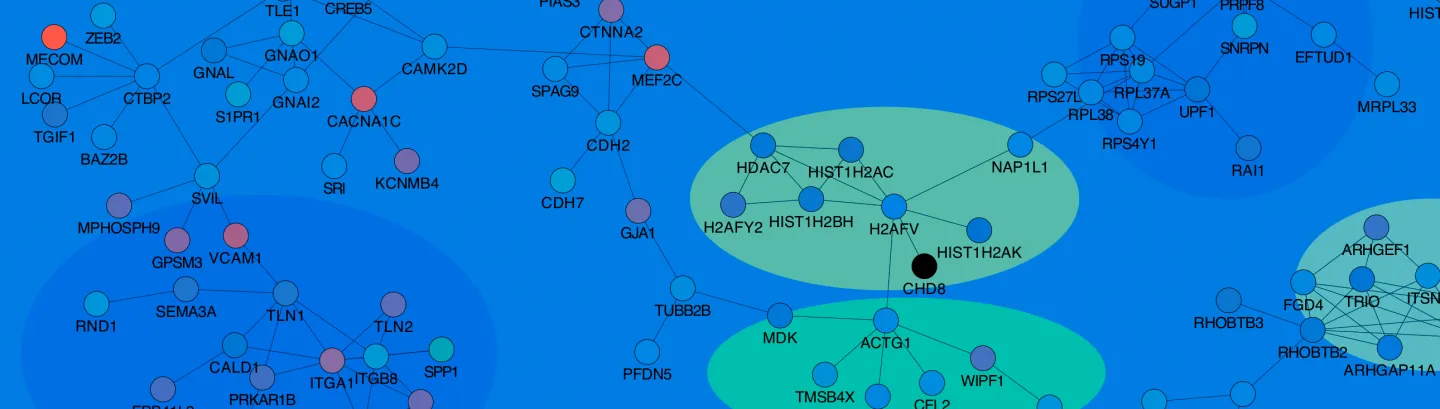
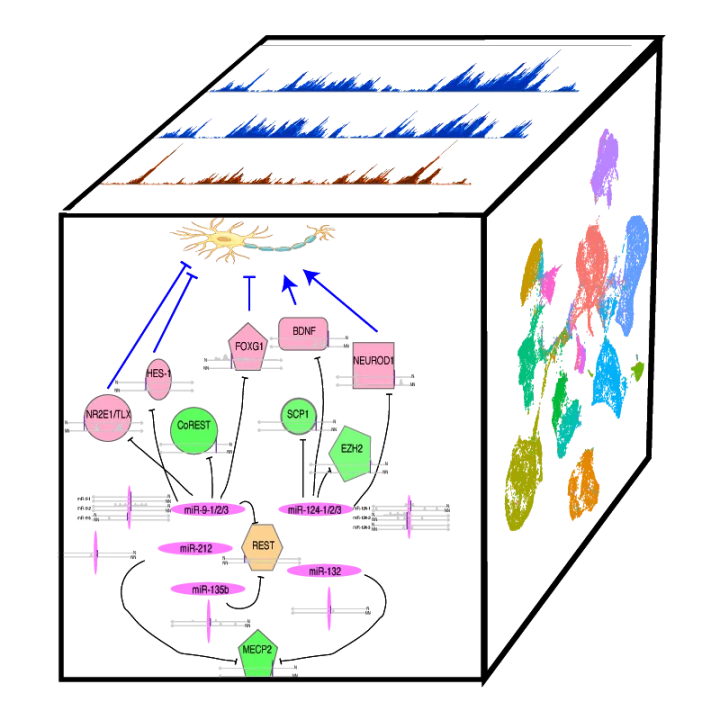
Decoding the human genome and other mammalian genomes to understand human diseases is our primary research interest, and comparative genomics and integrated analysis of functional genomics data are our major research approaches. In general, we are interested in developing and applying computational methods for exploiting patterns in high-throughput genomic and epigenomic data and for extracting biological information from experimental and computational bigdata. Since the lab was established in 2007, we have been developing bioinformatics approaches and software in studying the expression, regulation, and evolution of genes (both coding and non-coding) for human cardiac and nervous systems, especially those implicated in neuropsychiatric disorders and congenital heart diseases. We have substantial experiences in the analysis, interpretation, and integration of big experimental data from next-generation sequencing (e.g., ChIP-seq, ATAC-seq, RNA-seq and BS-seq) at bulk tissue and single cell levels, supported by > 200 preprints and publications. Using bioinformatics and in collaboration with experimental investigators, we want to understand how a single human genome is used for establishing and maintaining distinct cell lineages and how this important process is abnormally altered during disease development. Our research group focuses on multiple areas.

Lab Members
Our research team comprises graduate students and postdoctoral fellows representing a broad spectrum of cultural, ethnic, and educational backgrounds. Prior to joining the laboratory, some members received training as experimental biologists, whereas others were educated as data scientists with varying degrees of computational experience. A central mission of the lab is to cultivate and mentor its members to become a leading researcher in bioinformatics and genomics, supporting both their scientific growth and long-term career development.
Publications
- Misra R, Ferrena A, Zheng D. (2025) Facilitate integrated analysis of single cell multiomic data by binarizaing gene expression values. Nat Commu. 16:5763
- Ferrena A, Zheng XY, Jackson K, Hoang B, Morrow BE, Zheng D. (2024) scDAPP: a comprehensive single-cell transcriptomics analysis pipeline optimized for cross-group comparison. Nar Genom Bioinform. 6:lqaue134.
- Astorkia M, Lachman HM, Zheng D. (2022) Characterization of cell-cell communication in autistic brains with single-cell transcriptomics. J Neurodev Disord 14:29.
- Liu Y, Wang T, Zhou B, Zheng D. (2021) Robust integration of multiple single-cell RNA sequencing datasets using a single reference space. Nat Biotechnol. 39: 877-884