Bile Acids
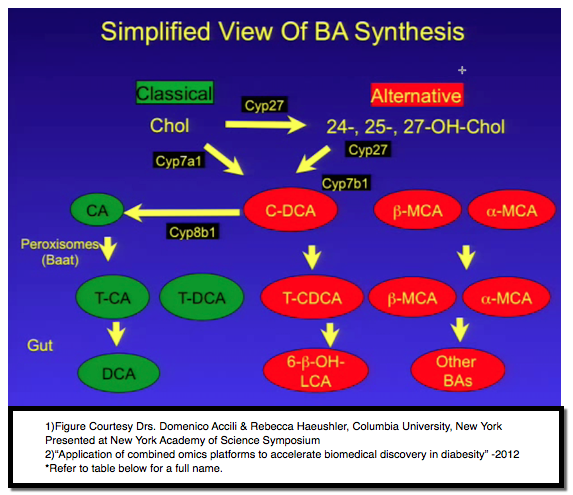
Bile acids are synthesized from cholesterol through both classical and alternative pathways. In the alternative pathway, the side chain oxidation of cholesterol precedes the steroid ring modifications, first yielding 24-, 25-, and 27-hydroxycholesterol metabolites, opposite to the process in the classical pathway. The alternative and classical pathway bile acids share the primary bile acid chenodeoxycholic acid, with 12-a-hydroxylation of chenodeoxycholic acid via CYP8B1to cholic acid. Modifications of bile acids can affect their properties and their ability to activate bile acid receptors. Dysregulation of bile acid synthesis can be seen in inborn errors of metabolism, insulin resistance, hepatocellular Ca and chronic ethanol consumption. Perturbations in the microbiome also affect bile acid pool size and composition (see references below). This panel surveys conditions of bile acid dysregulation.
For examples of this module’s utility for Core customers, see:
Bowden, J. A., Heckert, A., Ulmer, C. Z., Jones, C. M., Koelmel, J. P., Abdullah, L., Ahonen, L., Alnouti, Y., Armando, A., Asara, J. M., Bamba, T., Barr, J. R., Bergquist, J., Borchers, C. H., Brandsma, J., Breitkopf, S. B., Cajka, T., Cazenave-Gassiot, A., Checa, A., Cinel, M. A., Colas, R. A., Cremers, S., Dennis, E. A., Evans, J. E., Fauland, A., Fiehn, O., Gardner, M. S., Garrett, T. J., Gotlinger, K. H., Han, J., Huang, Y., Neo, A. H., Hyotylainen, T., Izumi, Y., Jiang, H., Jiang, H., Jiang, J., Kachman, M., Kiyonami, R., Klavins, K., Klose, C., Kofeler, H. C., Kolmert, J., Koal, T., Koster, G., Kuklenyik, Z., Kurland, I. J., Leadley, M., Lin, K., Maddipati, K. R., McDougall, D., Meikle, P. J., Mellett, N. A., Monnin, C., Moseley, M. A., Nandakumar, R., Oresic, M., Patterson, R. E., Peake, D., Pierce, J. S., Post, M., Postle, A. D., Pugh, R., Qui, Y., Quehenberger, O., Ramrup, P., Rees, J., Rembiesa, B., Reynaud, D., Roth, M. R., Sales, S., Schuhmann, K., Schwartzman, M. L., Serhan, C. N., Shevchenko, A., Somerville, S. E., John-Williams, L. S., Surma, M. A., Takeda, H., Thakare, R., Thompson, J. W., Torta, F., Triebl, A., Trotzmuller, M., Ubhayasekera, S. J. K., Vuckovic, D., Weir, J. M., Welti, R., Wenk, M. R., Wheelock, C. E., Yao, L., Yuan, M., Zhao, X. H. & Zhou, S. (2017) Harmonizing Lipidomics: NIST Interlaboratory Comparison Exercise for Lipidomics using Standard Reference Material 1950 Metabolites in Frozen Human Plasma, Journal of lipid research.
References:
- More detailed pathways see Bile acid Biosynthesis, Primary Bile acid Biosynthesis, Secondary Bile acid Biosynthesis
- Human insulin resistance is associated with increased plasma
levels of 12a-hydroxylated bile acids Haeusler RA, Astiarraga B, Camastra S, Accili D, Ferrannini E. Diabetes. 2013 Dec; 62(12):4184-91. doi: 10.2337/db13-0639.
Epub 2013 Jul 24
- Application of combined omics platforms to accelerate biomedical
discovery in diabesity. Kurland
IJ, Accili D, Burant C,
Fischer SM, Kahn BB, Newgard CB, Ramagiri S, Ronnett GV, Ryals JA, Sanders M,
Shambaugh J, Shockcor J, Gross SS. Ann N Y Acad Sci.
2013 May;1287:1-16. doi: 10.1111/nyas.12116. Epub 2013 May 9.
- Serum and urine metabolite profiling reveals potential
biomarkers of human hepatocellular carcinoma. Chen T, Xie G, Wang X, Fan J, Qiu Y, Zheng X, Qi X,
Cao Y, Su M, Wang X, Xu LX, Yen Y, Liu P, Jia W. Mol Cell
Proteomics. 2011 Jul; 10(7):M110.004945. doi: 10.1074/mcp.M110.004945.
Epub 2011 Apr 25. Erratum in: Mol Cell Proteomics. 2011 Nov; 10(11).
doi:10.1074/mcp.A110.004945.
- Alteration of bile acid metabolism in the rat induced by
chronic ethanol consumption.Xie G, Zhong
W, Li H, Li Q, Qiu Y, Zheng X, Chen H, Zhao X, Zhang S, Zhou Z, Zeisel SH, Jia
W. FASEB J. 2013 Sep;27(9):3583-93. doi:
10.1096/fj.13-231860. Epub 2013 May 24.
- Bile acids and the gut microbiome. Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS.Curr Opin Gastroenterol. 2014 May;30(3):332-8
< Module 4 | Module 6 >